22. Bayesian Model Comparison#
What alternatives do we have for AIC and BIC when we don’t have a maximum Liklihood?
import numpy as np
import pymc as pm
import arviz as az
import matplotlib.pyplot as plt
import seaborn as sns
sns.set_theme(context="talk", style="ticks", font="serif", color_codes=True)
np.random.seed(0)
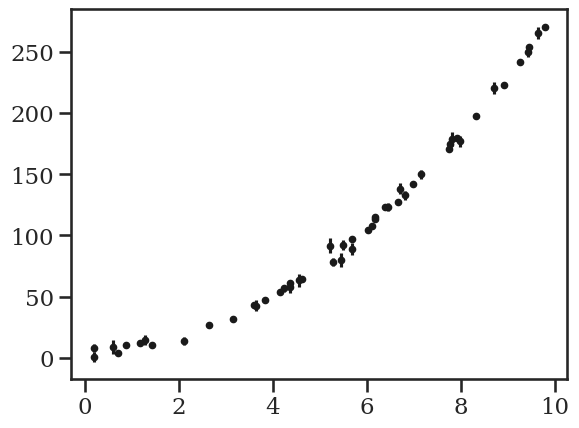
c_true = (5, 0.12, 11/4)
N = 50
x = np.sort(10*np.random.rand(N))
yerr = 1 + 5 * np.random.rand(N)
y = c_true[0] + c_true[1]*x + c_true[2]*x**2
y += yerr * np.random.randn(N)
plt.errorbar(x, y, yerr=yerr, fmt="k.");
# model is y = c0 + c1*x + c2*x^2 + noise
# build a model via a `with` context block.
with pm.Model() as model:
# define priors on polynomial coeffiencts and the added noise
c0 = pm.Normal('c0', 0, 20)
c1 = pm.Normal('c1', 0, 20)
c2 = pm.Normal('c2', 0, 20)
noise = pm.Exponential('noise', 0.1) #could be half-Normal or half-chauchy
# define observations: y = c0 + c1*x + c2*x^2
# link variables to data with `observed=y`
obs = pm.Normal('obs', c0 + c1*x + c2*x**2, noise, observed=y)
# run sampler
idata_quad = pm.sample(idata_kwargs={"log_likelihood": True})
idata_quad
Initializing NUTS using jitter+adapt_diag...
Multiprocess sampling (4 chains in 4 jobs)
NUTS: [c0, c1, c2, noise]
Sampling 4 chains for 1_000 tune and 1_000 draw iterations (4_000 + 4_000 draws total) took 1 seconds.
-
<xarray.Dataset> Size: 136kB Dimensions: (chain: 4, draw: 1000) Coordinates: * chain (chain) int64 32B 0 1 2 3 * draw (draw) int64 8kB 0 1 2 3 4 5 6 7 ... 993 994 995 996 997 998 999 Data variables: c0 (chain, draw) float64 32kB 5.283 5.152 4.72 ... 8.296 3.909 5.457 c1 (chain, draw) float64 32kB 0.3898 0.5193 0.1933 ... 1.067 0.8632 c2 (chain, draw) float64 32kB 2.746 2.73 2.781 ... 2.809 2.67 2.659 noise (chain, draw) float64 32kB 3.469 3.623 3.391 ... 3.345 3.842 4.016 Attributes: created_at: 2025-04-23T18:50:56.582996+00:00 arviz_version: 0.21.0 inference_library: pymc inference_library_version: 5.22.0 sampling_time: 1.480475902557373 tuning_steps: 1000 -
<xarray.Dataset> Size: 2MB Dimensions: (chain: 4, draw: 1000, obs_dim_0: 50) Coordinates: * chain (chain) int64 32B 0 1 2 3 * draw (draw) int64 8kB 0 1 2 3 4 5 6 7 ... 993 994 995 996 997 998 999 * obs_dim_0 (obs_dim_0) int64 400B 0 1 2 3 4 5 6 7 ... 43 44 45 46 47 48 49 Data variables: obs (chain, draw, obs_dim_0) float64 2MB -3.128 -2.431 ... -2.369 Attributes: created_at: 2025-04-23T18:50:56.698044+00:00 arviz_version: 0.21.0 inference_library: pymc inference_library_version: 5.22.0 -
<xarray.Dataset> Size: 496kB Dimensions: (chain: 4, draw: 1000) Coordinates: * chain (chain) int64 32B 0 1 2 3 * draw (draw) int64 8kB 0 1 2 3 4 5 ... 995 996 997 998 999 Data variables: (12/17) index_in_trajectory (chain, draw) int64 32kB -6 -5 -5 -8 ... -21 -21 -3 tree_depth (chain, draw) int64 32kB 4 4 5 5 6 5 ... 4 5 6 6 6 5 step_size_bar (chain, draw) float64 32kB 0.1268 0.1268 ... 0.129 step_size (chain, draw) float64 32kB 0.1536 0.1536 ... 0.163 energy (chain, draw) float64 32kB 148.4 147.9 ... 150.4 perf_counter_diff (chain, draw) float64 32kB 0.0003504 ... 0.0006941 ... ... largest_eigval (chain, draw) float64 32kB nan nan nan ... nan nan smallest_eigval (chain, draw) float64 32kB nan nan nan ... nan nan energy_error (chain, draw) float64 32kB 0.3499 ... 0.05073 perf_counter_start (chain, draw) float64 32kB 2.979e+04 ... 2.979e+04 diverging (chain, draw) bool 4kB False False ... False False process_time_diff (chain, draw) float64 32kB 0.000351 ... 0.000695 Attributes: created_at: 2025-04-23T18:50:56.592271+00:00 arviz_version: 0.21.0 inference_library: pymc inference_library_version: 5.22.0 sampling_time: 1.480475902557373 tuning_steps: 1000 -
<xarray.Dataset> Size: 800B Dimensions: (obs_dim_0: 50) Coordinates: * obs_dim_0 (obs_dim_0) int64 400B 0 1 2 3 4 5 6 7 ... 43 44 45 46 47 48 49 Data variables: obs (obs_dim_0) float64 400B 0.6327 8.013 8.837 ... 253.6 265.0 270.0 Attributes: created_at: 2025-04-23T18:50:56.594554+00:00 arviz_version: 0.21.0 inference_library: pymc inference_library_version: 5.22.0
# model is y = c0 + c1*x + noise
# build a model via a `with` context block.
with pm.Model() as model:
# define priors on polynomial coeffiencts and the added noise
c0 = pm.Normal('c0', 0, 20)
c1 = pm.Normal('c1', 0, 20)
noise = pm.Exponential('noise', 0.1) #could be half-Normal or half-chauchy
# define observations: y = c0 + c1*x + c2*x^2
# link variables to data with `observed=y`
obs = pm.Normal('obs', c0 + c1*x, noise, observed=y)
# run sampler
idata_linear = pm.sample(idata_kwargs={"log_likelihood": True})
Initializing NUTS using jitter+adapt_diag...
Multiprocess sampling (4 chains in 4 jobs)
NUTS: [c0, c1, noise]
Sampling 4 chains for 1_000 tune and 1_000 draw iterations (4_000 + 4_000 draws total) took 1 seconds.
# model is y = c0 + c1*x + c2*x^2 + c3*x^3 + noise
# build a model via a `with` context block.
with pm.Model() as model:
# define priors on polynomial coeffiencts and the added noise
c0 = pm.Normal('c0', 0, 20)
c1 = pm.Normal('c1', 0, 20)
c2 = pm.Normal('c2', 0, 20)
c3 = pm.Normal('c3', 0, 20)
noise = pm.Exponential('noise', 0.1) #could be half-Normal or half-chauchy
# define observations: y = c0 + c1*x + c2*x^2 + c3*x^3
# link variables to data with `observed=y`
obs = pm.Normal('obs', c0 + c1*x + c2*x**2 + c3*x**3, noise, observed=y)
# run sampler
idata_cubic = pm.sample(idata_kwargs={"log_likelihood": True})
Initializing NUTS using jitter+adapt_diag...
Multiprocess sampling (4 chains in 4 jobs)
NUTS: [c0, c1, c2, c3, noise]
Sampling 4 chains for 1_000 tune and 1_000 draw iterations (4_000 + 4_000 draws total) took 5 seconds.
There were 2 divergences after tuning. Increase `target_accept` or reparameterize.
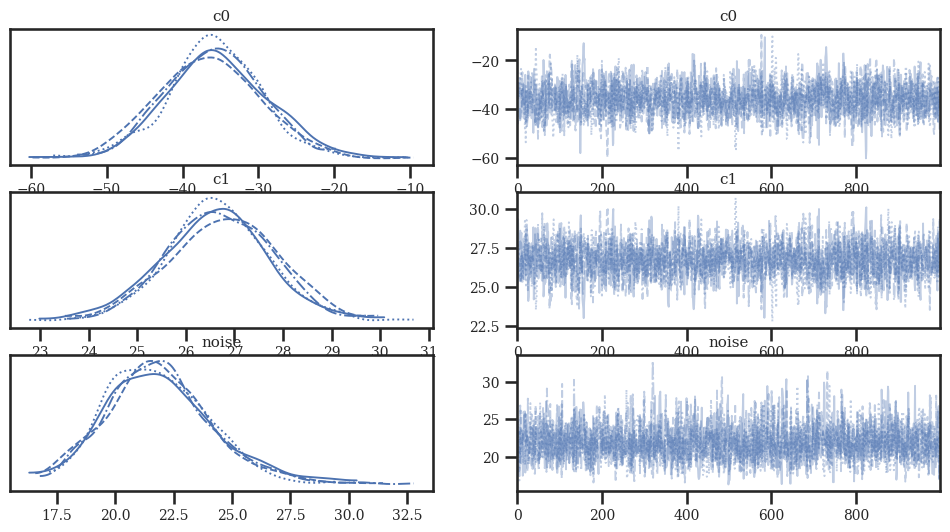
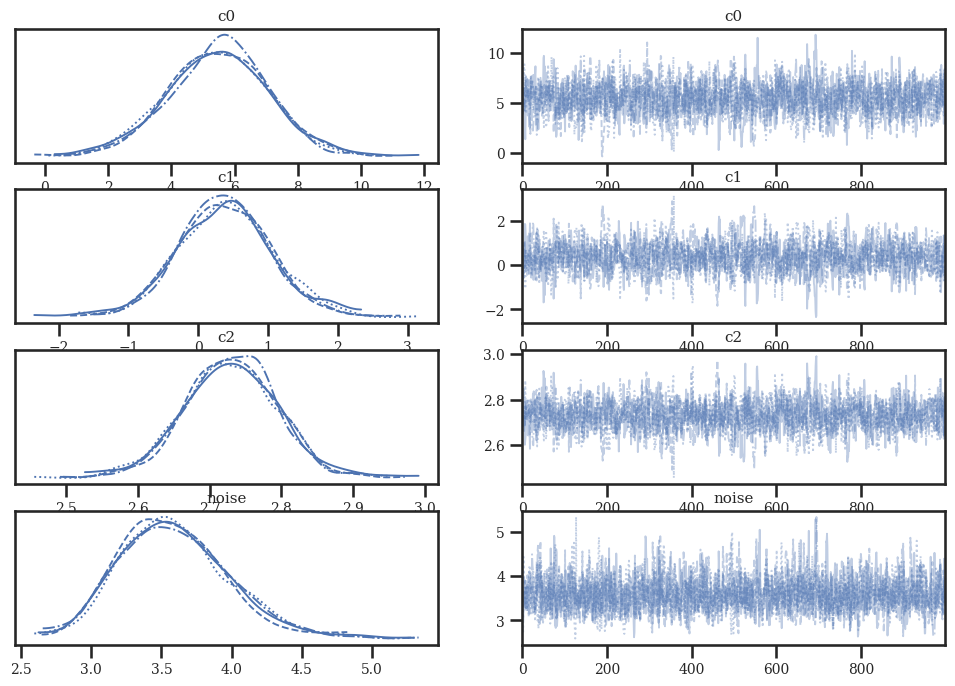
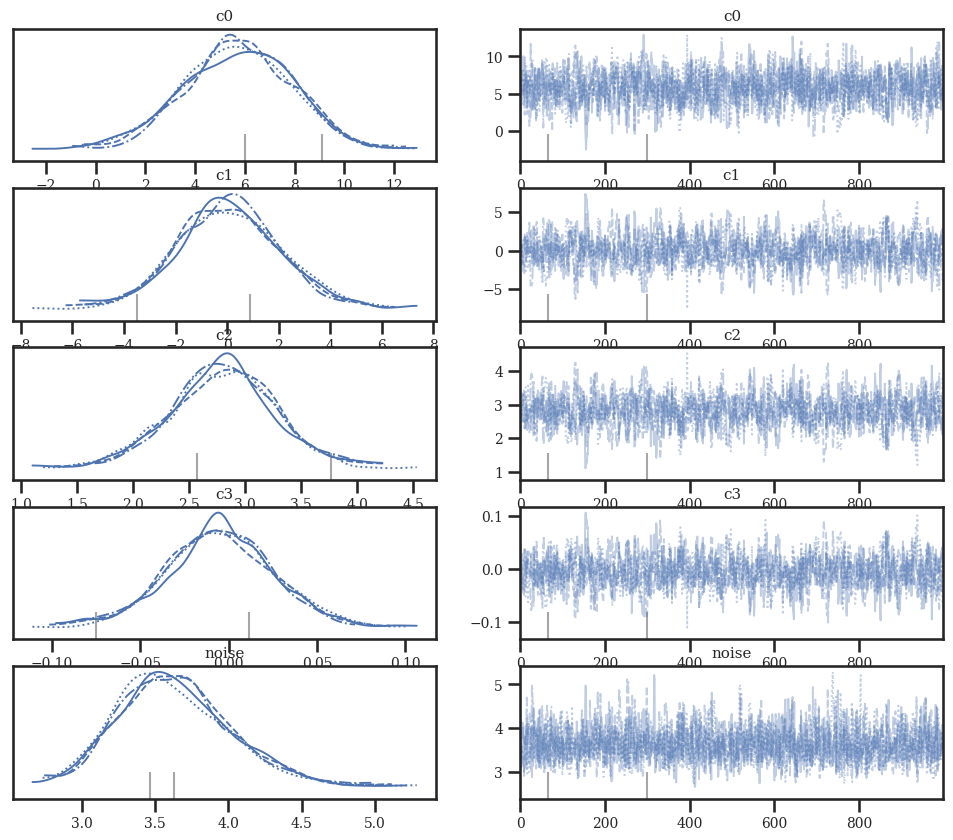
az.plot_trace(idata_linear)
az.plot_trace(idata_quad)
az.plot_trace(idata_cubic);
22.1. Cross Valication & Leave-one-out#
Gelman, Hill, & Vehtari section 11.8 describes the theory behind the following process
https://python.arviz.org/en/stable/api/generated/arviz.compare.html https://python.arviz.org/en/latest/api/generated/arviz.plot_compare.html
Note, you need idata_kwargs={"log_likelihood": True} in the pm.sample() function in order to save the likelihoods needed for the fast leave-one-out calcualtions described in Gelman 11.8.
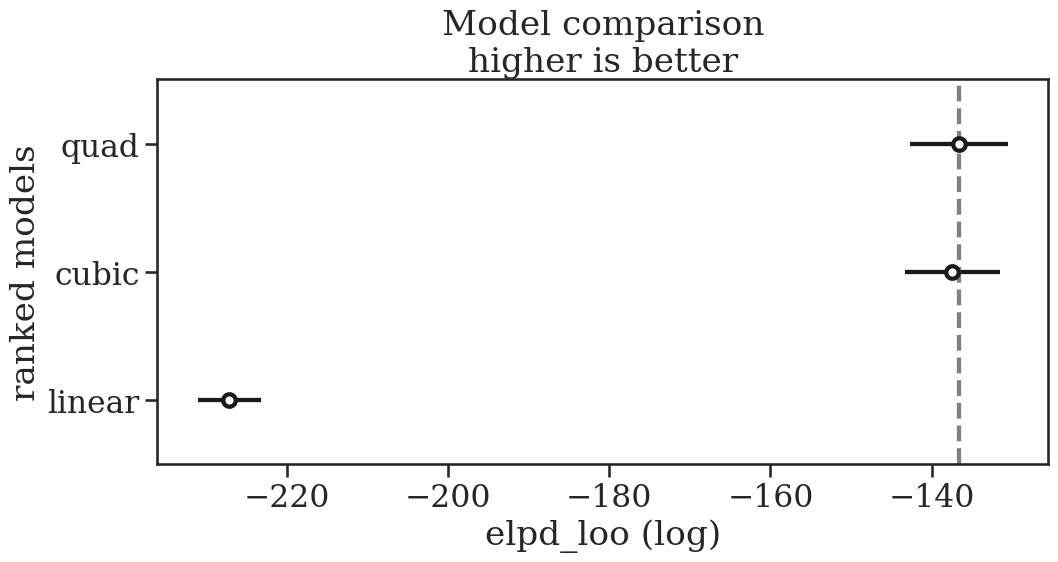
# Perform the Leave-one-out calcualtion
idata_linear = az.loo(idata_linear)
idata_quad = az.loo(idata_quad)
idata_cubic = az.loo(idata_cubic)
model_compare = az.compare(
{
"linear": idata_linear,
"quad": idata_quad,
"cubic": idata_cubic,
}
)
az.plot_compare(model_compare, figsize=(11.5, 5));
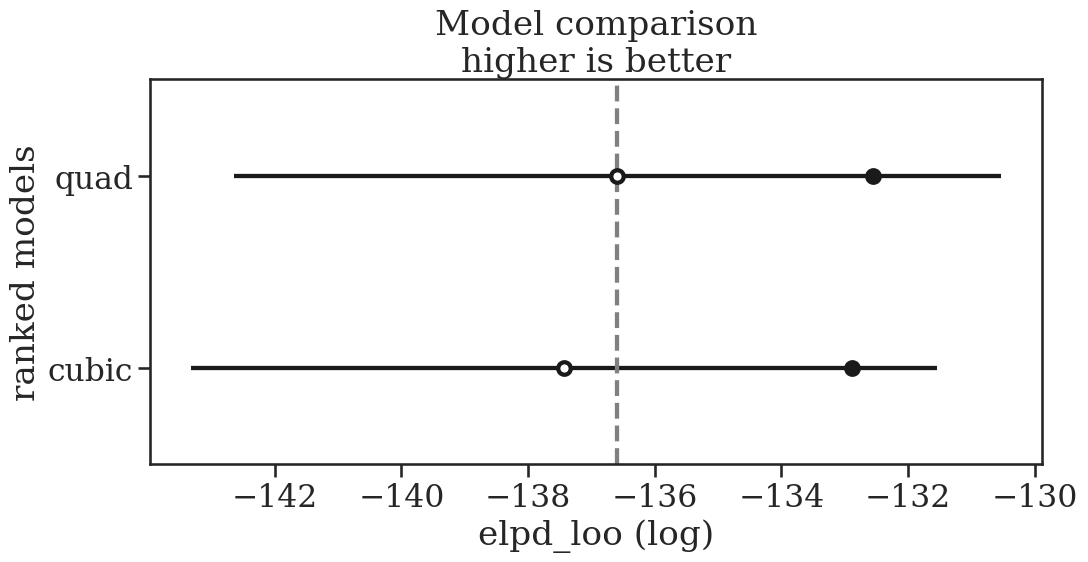
# Lets zoom in on quad and cubic
model_compare = az.compare(
{
"quad": idata_quad,
"cubic": idata_cubic,
}
)
az.plot_compare(model_compare, figsize=(11.5, 5), insample_dev=True);
22.2. Further Reading#
Gelman, Hill, Vehtari chapter 11
https://python.arviz.org/en/latest/api/generated/arviz.loo.html#arviz.loo
https://python.arviz.org/en/stable/api/generated/arviz.compare.html
https://python.arviz.org/en/latest/api/generated/arviz.plot_compare.html
https://www.pymc.io/projects/examples/en/latest/generalized_linear_models/GLM-model-selection.html
%load_ext watermark
%watermark -untzvm -iv -w
Last updated: Wed Apr 23 2025 13:53:53CDT
Python implementation: CPython
Python version : 3.11.12
IPython version : 9.1.0
Compiler : Clang 18.1.8
OS : Darwin
Release : 24.4.0
Machine : arm64
Processor : arm
CPU cores : 12
Architecture: 64bit
pandas : 2.2.3
numpy : 1.25.2
rich : 14.0.0
pymc : 5.22.0
seaborn : 0.13.2
matplotlib: 3.10.1
arviz : 0.21.0
Watermark: 2.5.0